Abstract
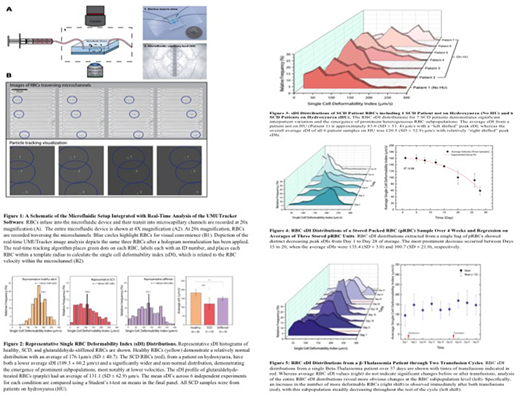
Background: Studying individual red blood cells (RBCs) is critical to understanding hematologic diseases, as pathology often originates at the single-cell level. Measuring biophysical properties, such as deformability, of RBCs is also clinically important as RBC deformability is pathologically altered in numerous disease states including sickle cell disease (SCD), beta-thalassemia major (β-thal), and the "storage lesion" that occurs over time in stored samples of otherwise normal RBCs1-3. While ektacytometry-based systems measure RBC deformability, these assays do not have single-cell resolution4. As evidenced by flow cytometry, single cell measurements are important to detect pathologic cellular subpopulations in hematologic diseases. Recently, microfluidic devices that model the microvascular environment have enabled single RBC deformability measurements, but truly high-throughput measurements have remained elusive due to technical issues such as image processing inaccuracy or aberrant signaling when reconstructing RBC velocity profiles.
Methods: To that end, here we introduce a novel integration of a microfluidic device coupled with our UMUTracker, an innovative MATLAB-based automated particle tracking program that is run on a standard desktop computer5. This innovative pairing of the two technologies results in high-throughput velocity tracking of single RBCs through a microfluidic model of a capillary bed that translates to a single deformability index (sDI) for each RBC (Fig 1). To demonstrate the sDI heterogeneity of RBCs across different conditions, data was obtained from healthy volunteers, SCD and β-thal patients, and stored packed RBCs.
Results: sDI distribution curves were obtained for healthy RBCs and glutaraldehyde-stiffened RBCs (as positive controls) in which the latter resulted in an expected "left shift" and decrease in the mean sDI (Fig 2). Interestingly, while the mean sDI for SCD RBCs was also expectedly decreased, the sDI distribution was clearly non-normal, indicating the existence of heterogeneous RBC subpopulations with different deformabilities. These heterogeneous sDI distributions were observed in multiple SCD patients, including those on hydroxyurea (HU) and not, although patients on HU exhibited RBCs subpopulations with relatively higher sDIs (Fig 3). While stored RBC samples showed an expected drop in mean sDI over time, our system detected marked shifts in peak sDI with high temporal resolution over only 4 day intervals (Fig 4). Finally, each time a β-thal patient was transfused, the peak sDI shifted to the right and then gradually decreased over the course of the transfusion cycle, while the mean sDI exhibited minimal change over time (Fig 5).
Conclusions: Our novel combined microfluidic/portable image analysis system demonstrates the high-throughput capability to detect distinct RBC subpopulations, at the single cell level, of different deformabilities in SCD, β-thal, and aging stored RBCs. This heterogeneity indicates that, in these disease states, RBC deformability cannot be fully characterized with mean or bulk biophysical measurements such as those obtained with ektacytometry. Ongoing studies will determine how changes in sDI profiles are associated with clinical events and different therapies as well as the biological significance of these RBC subpopulations with varied deformabilities and the underlying mechanisms for these differences.
References:
García-Roa M, del Carmen Vicente-Ayuso M, Bobes AM, et al. Red blood cell storage time and transfusion: current practice, concerns and future perspectives. Blood Transfusion. 2017;15(3):222-231.
Li X, Dao M, Lykotrafitis G, Karniadakis GE. Biomechanics and biorheology of red blood cells in sickle cell anemia. Journal of biomechanics. 2017;50:34-41.
Mangalani M, Lokeshwar MR, Banerjee R, Nageswari K, Puniyani RR. Hemorheological changes in blood transfusion-treated beta thalassemia major patients. Clin Hemorheol Microcirc. 1998;18(2-3):99-102.
Rabai M, Detterich JA, Wenby RB, et al. Deformability analysis of sickle blood using ektacytometry. Biorheology. 2014;51(0):159-170.
Zhang H, Stangner T, Wiklund K, Rodriguez A, Andersson M. UmUTracker: A versatile MATLAB program for automated particle tracking of 2D light microscopy or 3D digital holography data. Computer Physics Communications. 2017;219:390-399.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal